Enhancing the root microbiome in canola
Date: May 31, 2021
Term: 3 years
Status: Completed
Researcher(s): Chantel Hamel, Agriculture and Agri-food Canada; Yantai Gan, Agriculture and Agri-food Canada; Luke Bainard, AAFC; Marc St. Arnaud, Jardin botanique de Montréal; Mohamed Hijri, University of Montreal; Chih-Ying Lay, University of Montreal
SaskCanola Investment: $66,633
Total Project Cost: $547,551
Funding Partners: NSERC, ACPC, MCGA
Grower Benefits
Biodiverse agricultural systems improve agricultural productivity and climate management.
Brassicaceae host plants were consistently significant in structuring the bacterial communities. Canola microbiomes were distinguished between roots and rhizosphere and were significantly different from those of the reference crops (pea and wheat).
Seeding density and plant nutrition modified the abundance of other bacterial and fungal taxa forming the core microbiomes of canola that are expected to impact crop growth.
Diversified pulse-oilseed cropping sequences are highly desirable to achieve high N₂ fixation and retention and minimize nitrogen loss. Lentil substantially increased biological N₂ fixation and reduced denitrification in the following oilseed crops.
Project Summary
Researchers wanted to continue the work to learn more about root microbiomes, identify the core microbiome in canola and in turn, improve fertilizer efficency in canola. This project had two main goals: a) to assess the composition of the canola root microbiome, and validate a list of reliable microbial taxa; and b) to study crop rotation systems and which best support the establishment of beneficial root microbiomes in canola and in other rotation crops. In order to reach these main goals, the project was further split into five smaller objectives.
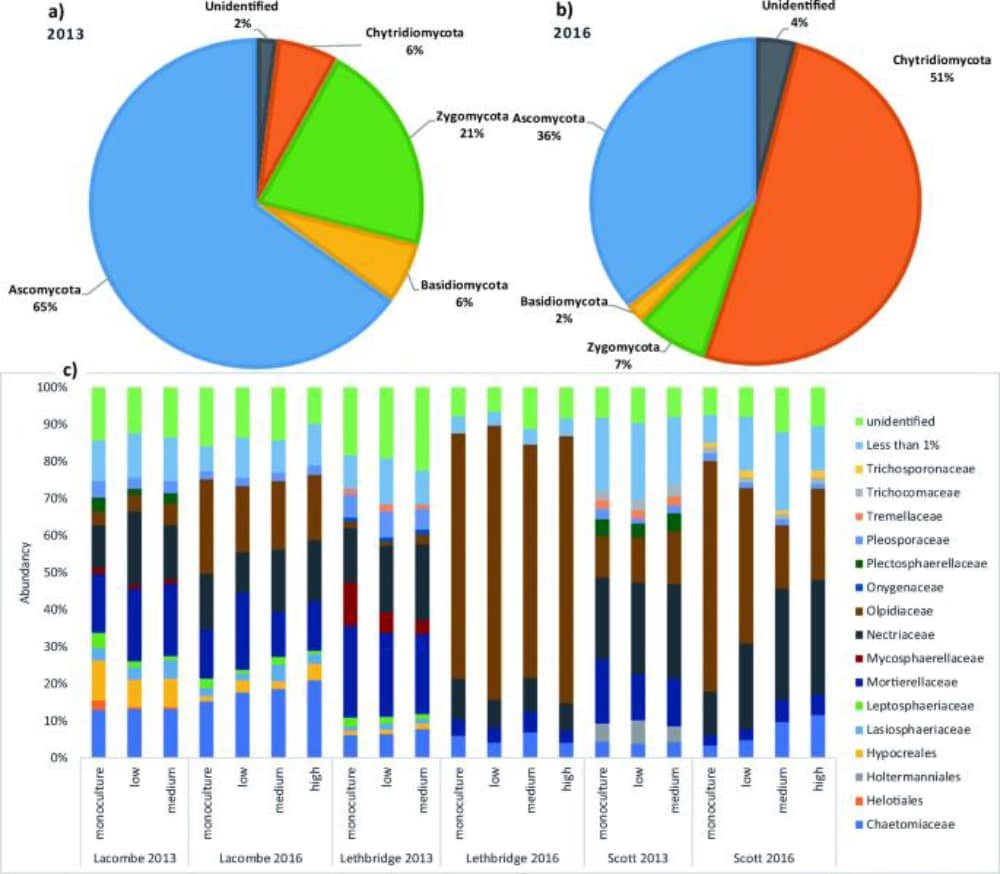
The first objective was to describe the canola root microbiome as influenced by different crop rotation systems over time, on a Brown and Black chernozem soil. Researchers wanted to determine if canola has a core root microbiome and if it differs from pea and wheat in a complex rotation system. Researchers also looked at whether selected agronomic practices can modify the canola microbiome and if any could be associated with enhanced yield. Roots and rhizosphere soil were harvested at the flowering stage of canola, and DNA from roots and soil was isolated separately. It was determined that the microbiome of the roots and rhizosphere of canola was consistently different from that of wheat and pea. The plant-growth-promoting rhizobacteria found included Amycolatopsis sp., Serratia proteamaculans, Pedobacter sp., Arthrobacter sp., Stenotrophomonas sp., Fusarium merismoides, and Fusicolla sp. The presence of this core microbiome correlated positively with canola yield. Individual crop species significantly influenced bacterial and fungal assemblages within their roots, while higher seeding density and nutrient input did not significantly alter the composistion of bacteria, fungi or archaea within canola roots. A known pathogen of members of the Brassicaceae family, Olpidium brassicae, was reduced in roots of canola planted at a higher seeding density. The results of this study suggests that seeding density and plant nutrition modified the abundance of other bacterial and fungal taxa forming the core microbiomes of canola that are expected to impact crop growth.
The second and third objectives were studied together: a) to identify the rotation with the best efficiency of nitrogen (N) cycling in the canola rhizosphere by quantifying the expression of genes involved in the processes of biological N₂ fixation, nitrification and denitrification in canola rhizosphere, and b) to identify the root microbiome taxonomic profiles related to efficent nitrogen use by canola. Researchers assessed N-cycling gene expression patterns in the root and rhizosphere microbiomes of five oilseed crops (Brassica napus, Brassica carinata, Brassica juncea, Sinapsis alba and Camelina sativa) as influenced by three different 2-year crop rotations. The first phase consisted of fallow, lentil or wheat and the second phase consisted of one of the oilseed crops. Expression of some of the bacterial nitrification and denitrification-related genes showed that the microbiome of Ethiopian mustard (Brassica carinata) had the lowest and the microbiome of camelina had the highest potential for nitrogen loss. Brassica carinata showed the best performance with the highest yield and lowest impact on greehouse gas (GHG) emissions, while Camelina sativa was the opposite, with lower yield and higher denitrification potential. As a preceding crop for oilseeds, lentil could help to increase N₂ fixation, decrease nitrogen fertilization application while reducing the agricultural impact on the environment. Alternatively, wheat was a poor performer for sustaining oilseed production and could result in nitrogen loss and potentially higher GHG emissions compared to lentil.
The fourth objective was to evaluate the potential of canola root microbiomes to provide canola with tolerance to abiotic stress and pathogen pressure. The impact of the canola-cereals-pea rotation systems with different crop intensities (frequency of each crop in rotation) was tested on crop productivity, arbuscular mycorrhizal (AM) fungal diversity and structure within the roots and rhizosphere of each crop as well as the relationships between specific AM fungal microbiome members and crop productivity. Three rotation systems (intensifying canola, cereals or pulse over four years) were tested and the root and rhizosphere microbiomes were sampled for each rotation phase at two growth stages. Increasing the frequency of canola in a four year rotation did not reduce the productivity of the other crops or translate into reduced biodiversity of AM fungi in the roots or rhizospheres, except for in canola itself. Crop and cropping systems did alter the AM fungal community structure in both roots and rhizospheric environments of the plants with positive or negative correlations with crop productivity. This suggests that a simple modification of the cropping system could be used to manipulate root or plant microbiomes to improve crop productivity without increasing the amount of inputs for crop production.
Variation in taxonomic profiles (of fungal microbiome of the canola rhizosphere) is characterized by an increase in the abundance of the Olpidiaeae in the phylum Chytridiomycota in 2016. Fungal families also varied with site, crop diversification level, and year (c).
The final objective was to correlate the changes in microbial composition with plant performance and rotational practices in order to improve understanding of the interactions between microbes and the plants. Rhizosphere microbes influence each other while forming complex webs of interactions that may determine plant success. Identifying the key factors that make up the fungal microbiome of the rhizosphere will help in optimizing plant production. Results indicate that crop diversification has a significant effect on the structure of the rhizosphere fungal community, but not on fungal diversity. The researchers were also able to discover and describe a canola core microbiome with an abundance of Olipidium brassicae (Figure 1). The researchers also identified four hub taxa, however none of these belonged to the core microbiome or eco-microbiome for each year of sampling.
Full Report PDF: Enhancing the root microbiome in canola